Artificial Intelligence for Life in Space
Space biosciences research advances safe human space travel. The AI4LS team builds advanced computational frameworks using machine learning, deep learning, and artificial intelligence to model, predict, and mitigate spaceflight risks.
How can we help you advance your research?
Focus Areas
 Large Language Models and Foundation ModelsWe develop and deploy large language models to revolutionize real-time health decision-making during long-duration spaceflights. These models also address challenges on Earth, such as organizing and standardizing unstructured data for improved findability, accessibility, interoperability, and reproducibility (read more about FAIR data here). |  Knowledge GraphsIn collaboration with the SPOKE biomedical knowledge graph (spoke.ucsf.edu) and the National Science Foundation’s Proto-OKN initiative, we apply graph representation learning to advance our understanding of space biology. |
 Digital TwinsOriginally pioneered by NASA during the Apollo 13 mission, digital twins are now being trained to predict biological system responses to spaceflight stressors, enabling precision health interventions and system optimization. |  Causal InferenceTo uncover causal relationships within complex biological systems, we apply cutting-edge machine learning techniques in causal inference, enhancing our ability to predict and mitigate biological risks. |
 Multi-Modal Data IntegrationBy integrating diverse experimental datasets from the NASA Open Science Data Repository, we develop innovative methods to merge and analyze multi-modal data, unlocking new perspectives in space biology research. |  Federated LearningTo ensure privacy and reduce the need for extensive data downlink, we utilize federated learning and decentralized training to deploy predictive models both on Earth and in space. |
Projects
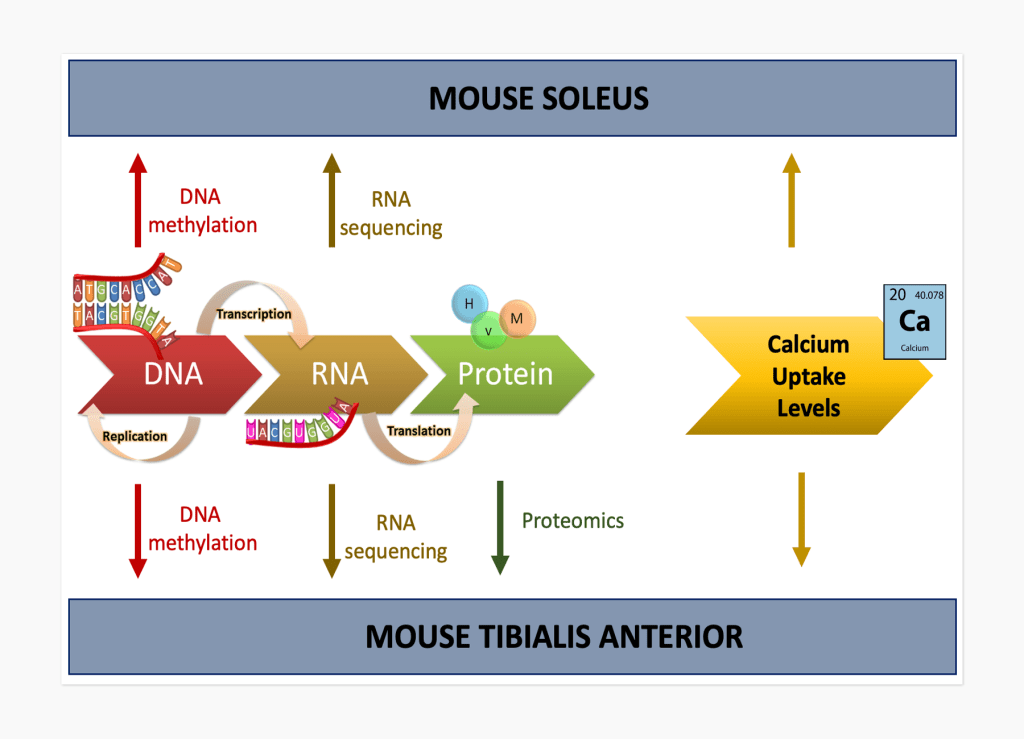
Multi-Modal Data and Explainable AI
Modeling the complex interplay between cellular networks and resulting physiological conditions requires integration of multi-modal data types. In Li et al npj Microgravity 2023, we leverage graph-based explainable AI through AbzuAI QLattice to provide transparent characterization of the relationship between biological features. We integrate multi-modal biological data (gene expression, protein expression, genomic methylation, and calcium uptake levels) from spaceflown mouse muscle to identify the relationships between molecular features to predict the changes in calcium uptake in muscle. In future work, we plan to develop advanced AI-based methods for integrating multi-modal complex biological datasets.
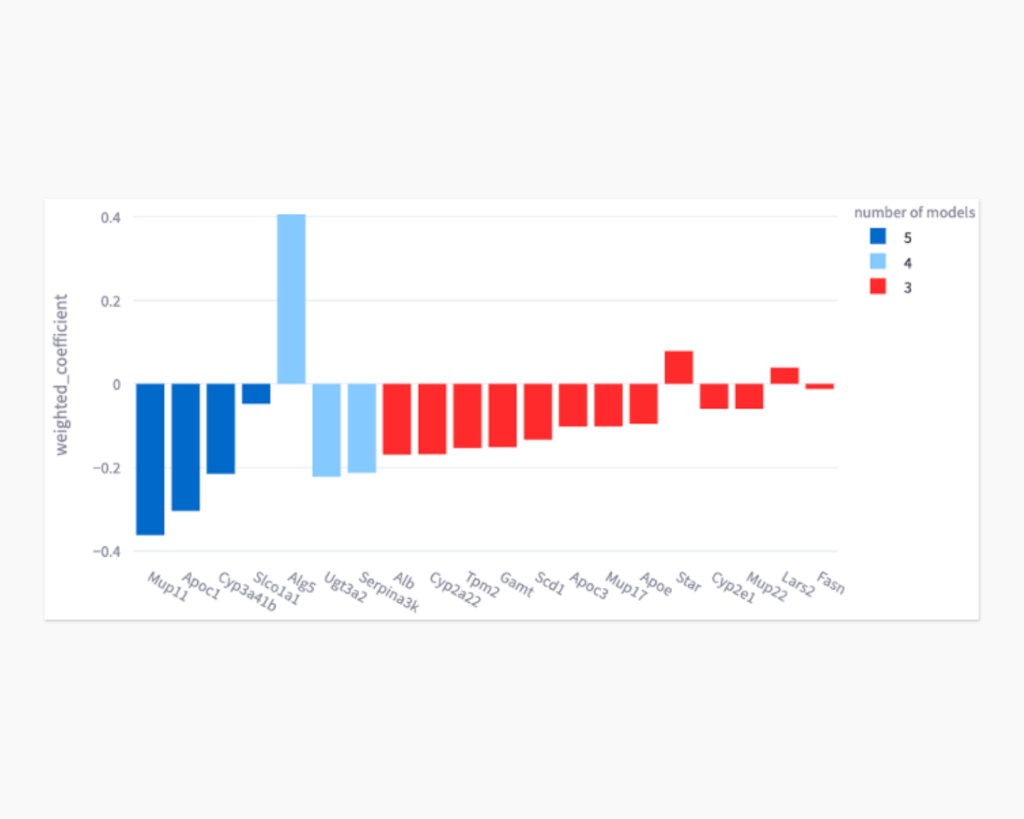
Causal Inference
Understanding causal relationships is essential for biomedical applications. We leverage an ensemble of causal inference machine learning methods (CRISP) to identify genes whose expression is potentially causal of increased fat content in liver from spaceflown mice. Currently, we are working with a Crowdsourcing Contenders award to develop a version of CRISP that can identify causal features in both tabular and bio-imagery data.
AI/ML Space Biology Certification Program
We provide open-source, no-cost, zero-prerequisite, virtual and asynchronous training in AI/ML for space biology. Enroll here: https://canvas.instructure.com/enroll/8JYKD7
Guided Transfer Learning
Collaboration with Robots Go Mental – Guided Transfer Learning (GTL) improves on traditional transfer learning by teaching the neural network “how” to learn through inductive biases that inform how the network learns in the future. We use GTL to achieve high classification performance in the high-dimensionality, low-sample-size settings intrinsic to space biology. Current and future work will focus on expanding GTL to few- and zero-shot learning.
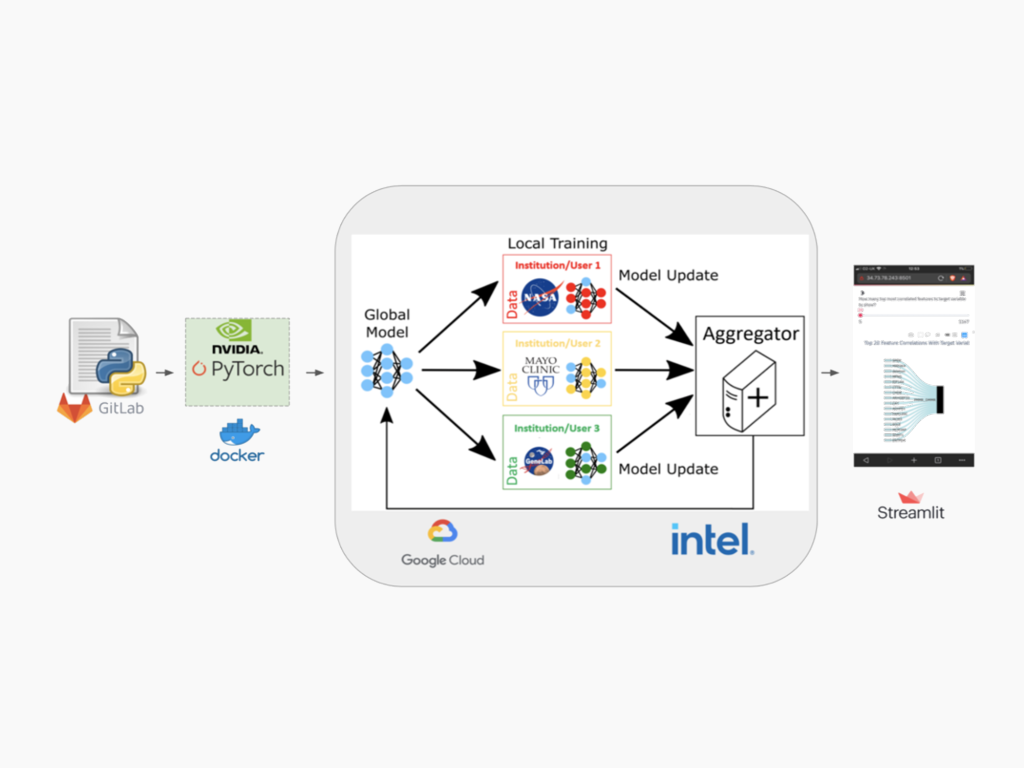
Federated Learning
Collaboration with Frontier Development Lab and Intel – The 2021 Frontier Development Lab Astronaut Health team, co-funded by NASA and Intel, developed an approach for federated learning to train predictive models with data on physically separated nodes. This approach enables model training in settings with low compute such as spaceflight, or settings with privacy concerns such as astronaut health. We leverage this approach for training and inference with the first federated machine learning model using data in space on the International Space Station and data held on Earth.
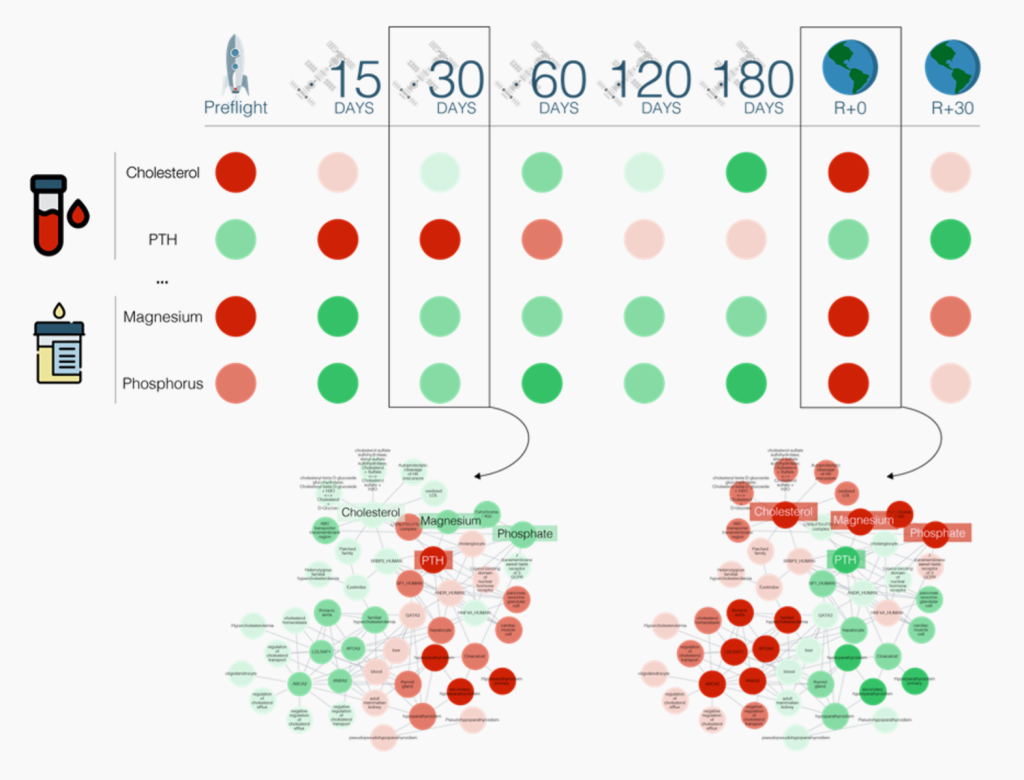
Knowledge Graphs
Collaboration with Baranzini Lab at UCSF – Leveraging multi-omics data from NASA GeneLab and integrating it with the Scalable Precision Medicine Oriented Knowledge Engine (SPOKE), the study below marks a step forward in identifying critical physiological changes and symptoms linked to spaceflight. SPOKE is being integrated into OSDR, creating a GeneLab-SPOKE fabric. Building on this approach, we aim to generalize the use of knowledge graphs (KGs) for monitoring and mitigating potential health risks for astronauts. By combining pre-, in-flight, and post-flight biomarker data (cholesterol, PTH, magnesium, phosphorus, etc.) from Earth and space, one should be able to refine actionable countermeasures to ensure astronaut health and safety in future missions.
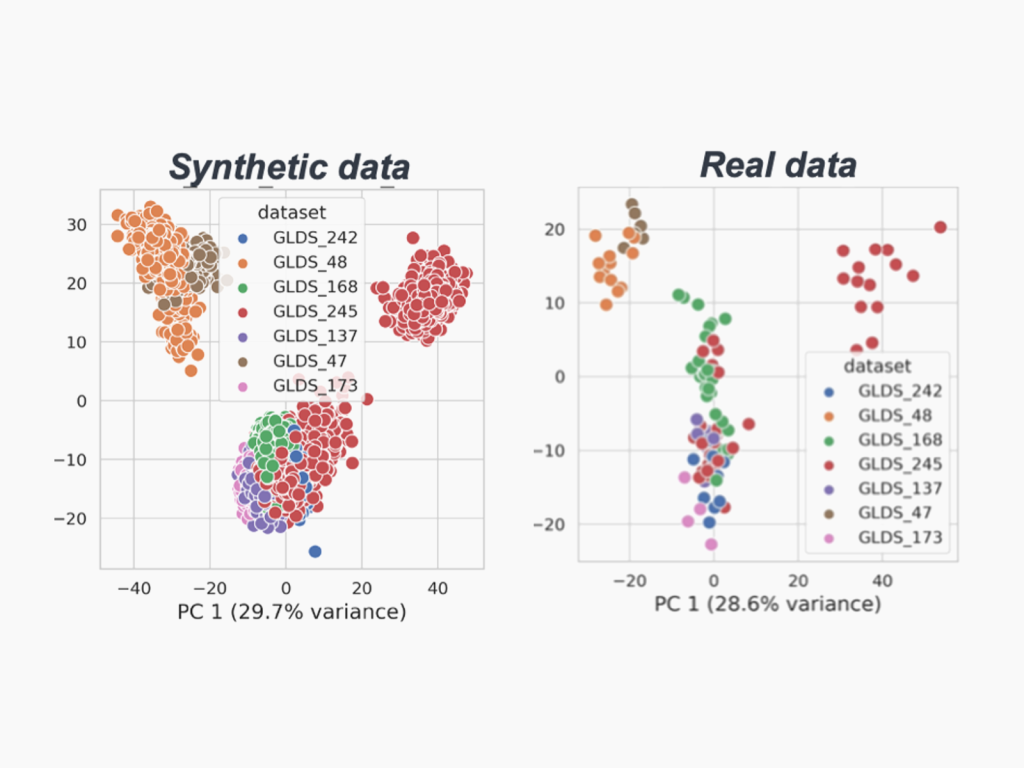
Realistic Synthetic Data Generation
Leveraging the Generative Adversarial Network (GAN), we develop realistic synthetic RNA sequencing gene expression data to amplify the signals in tiny space biological datasets. Current and future work focuses on evaluating additional architectures such as the Conditional Variational Autoencoder (CVAE).
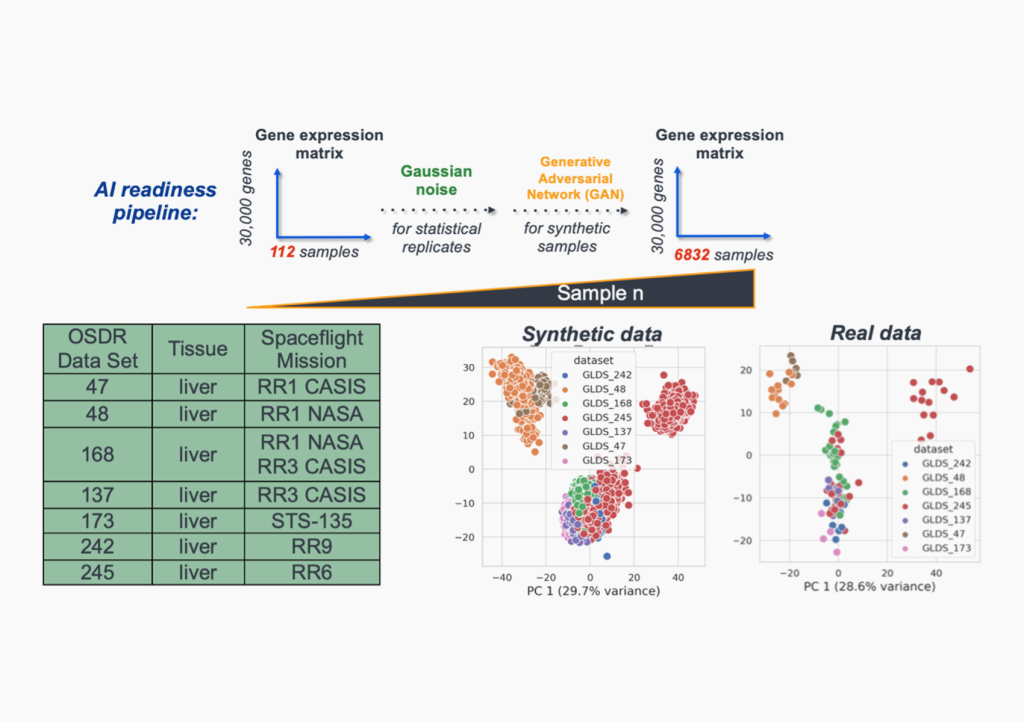
NASA Biological and Physical Sciences RNA Sequencing Benchmark Dataset
Supported by the NASA Science Mission Directorate, we publish a standardized, AI-ready RNA sequencing dataset for AI/ML benchmarking, training, and data challenges.
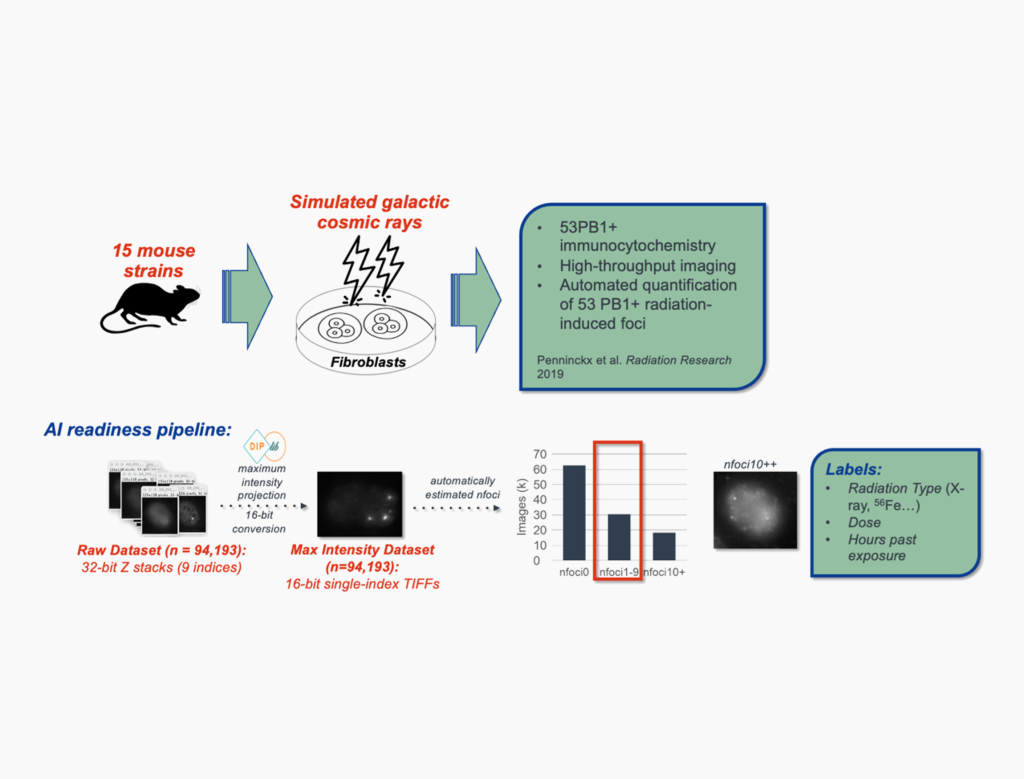
NASA Biological and Physical Sciences Microscopy Benchmark Dataset
Supported by the NASA Science Mission Directorate, we publish a standardized, AI-ready fluorescence microscopy dataset for AI/ML benchmarking, training, and data challenges.
Current Team Members
Previous Team Members
Konstantinos Adamopoulos
Sylvain Costes
Kayvon Crenshaw
Riya Desai
Brian Evarts
Sarah Golts
Adrienne Hoarfrost
Liam Johnson
Alice Joyeux
Kevin Li
Makenna Myrick
Charlotte Nelson
Dillon Nishigaya
Phani Paladugu
Sonali Patel
Lauren Sanders
Anmol Seth
Contact us!
Walter Alvarado (walter.alvarado@nasa.gov), James Casaletto (james.a.casaletto@nasa.gov)
Connect with us via the Open Science AI/ML Analysis Working Group (AWG)!
Current AWG Projects:
DNA Damage, Blood, Imaging/ML
Digital Twin Retina
Causal Inference
AI-Readiness
Anomaly Detection, Environmental Telemetry
AI-driven AOPs
Get certified in AI/ML for space biology! Sign up here for the asynchronous course: canvas.instructure.com/enroll/8JYKD7
Relevant Publications
- Casaletto JA, Zhao T, Yeung J, Lee A, Ansari A, Fry A, Mishra A, Raj A, Sun K, Lendahl S, Guan W, Cline MS, Costes SV. Machine Learning Ensemble Investigates Age in the Transcriptomic Response to Spaceflight in Murine Mammary Tissue: Observational Study. JMIRx Bio. 2026;4:e73041. doi:10.2196/73041
- Casaletto JA, Foley P, Fernandez M, Sanders LM, Scott RT, Ranjan S, Jain S, Haynes N, Boerma M, Costes SV, Mackintosh G. Bridging Earth and Space: A Flexible and Resilient Federated Learning Framework Deployed on the International Space Station. bioRxiv. 2025. doi:10.1101/2025.01.14.633017
- Casaletto JA, Scott RT, Myrick M, Adamopoulos K, Scheibenreif L, Patel SA, Soboczenski F, Hoarfrost A, Sanders LM, Costes SV. Analyzing the relationship between gene expression and phenotype in space-flown mice using a causal inference machine learning ensemble. Sci Rep. 2025;15:2363. doi:10.1038/s41598-024-81394-y
- Pantha, N. et al. Feature selection in high-dimensional space with applications to gene expression data. in SoutheastCon 2024 vol. 6 6–15 (IEEE, 2024).
- Li K, Desai R, Scott RT, Steele JR, Machado M, Demharter S, et al. Explainable machine learning identifies multi-omics signatures of muscle response to spaceflight in mice. NPJ Microgravity. 2023;9: 90. doi:10.1038/s41526-023-00337-5
- Nelson, C.A.; Acuna, A.U.; Paul, A.M.; Scott, R.T.; Butte, A.J.; Cekanaviciute, E.; Baranzini, S.E.; Costes, S.V. Knowledge Network Embedding of Transcriptomic Data from Spaceflown Mice Uncovers Signs and Symptoms Associated with Terrestrial Diseases. Life 2021, 11, 42. https://doi.org/10.3390/life11010042
- Li K, Nikolić D, Nikolić V, Andrić D, Sanders LM, Costes SV. Using Guided Transfer Learning to Predispose AI Agent to Learn Efficiently from Small RNA-sequencing Datasets. arXiv [q-bio.GN]. 2023. Available: http://arxiv.org/abs/2311.12045
- Sanders LM, Scott RT, Yang JH, Qutub AA, Martin HG, Berrios DC, et al. Biological Research and Self-Driving Labs in Deep Space supported by Artificial Intelligence. Nature Machine Intelligence. 2023.
- Scott RT, Sanders LM, Antonsen EL, Hastings JJA, Park S-M, Mackintosh G, et al. Biomonitoring and Precision Health in Deep Space supported by Artificial Intelligence. Nature Machine Intelligence. 2023.
- Duckworth P, O’Donoghue O, Scheibenreif L, Ughi G, Hoarfrost A, Budd S, et al. Federated causal inference for out-of-distribution generalization in predicting physiological effects of radiation exposure. Frontier Development Lab Technical Memorandum; 2021 Aug. Available: https://drive.google.com/file/d/1xM8USgeYs-NNbB83JSUl4EnfMG09W1Se/view
- Budd S, Blaas A, Hoarfrost A, Khezeli K, D’Silva K, Soboczenski F, et al. Prototyping CRISP: A Causal Relation and Inference Search Platform applied to Colorectal Cancer Data. 2021 IEEE 3rd Global Conference on Life Sciences and Technologies (LifeTech). ieeexplore.ieee.org; 2021. pp. 517–521. doi:10.1109/LifeTech52111.2021.9391819